Mark annotation #720
Unanswered
dorinemerlat
asked this question in
Q&A
Replies: 1 comment 1 reply
-
|
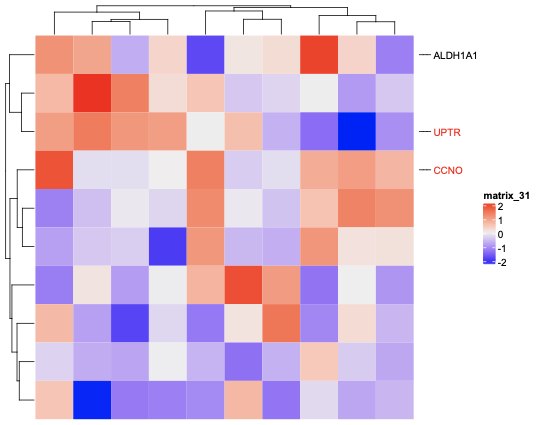
You can set annot = rowAnnotation(foo = anno_mark(
at = c(1, 4, 8),
labels = c("ALDH1A1", "UPTR", "CCNO"),
labels_gp = gpar(col = c("black", "red", "red"), fontsize = 10)))
Heatmap(matrix(rnorm(100), 10)) + annot |
Beta Was this translation helpful? Give feedback.
1 reply
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
-
Hi, I have a little issue with the mark annotations. I can add them to my heatmap but I would like to use two different colors following the fact that the genes are known or not. To add them I used that :
To obtain two different colors for annotation I thought I would use the list() in the different fields (like for the other annotation types), but it didn't work (error :
atshould be numeric row index corresponding to the matrix) :Is there any other way to put the annotations in different colors ?
Thanks :)
Beta Was this translation helpful? Give feedback.
All reactions