VariantAInalyser is a comprehensive platform that combines traditional bioinformatics with generative AI to streamline genomic variant analysis from VCF files. It eliminates the need to juggle multiple specialised tools by providing an all-in-one solution for variant analysis, interpretation, and reporting.
👩🏻🔬 The Challenge
Genomic variants are essential biomarkers for understanding diseases, drug responses, and creating personalised treatment plans. However, traditional analysis workflows force researchers to:
- Switch between multiple disconnected tools (VCF parsers, annotation software, database interfaces)
- Master different user interfaces and data formats
- Manually integrate results across platforms
- Spend valuable time on technical tasks rather than interpretation
This fragmented approach creates inefficiencies, increases the potential for errors, and significantly extends analysis time.
💡The Solution: VariantAInalyser!
VariantAInalyser revolutionises genomic analysis by unifying the entire workflow in a single, intuitive interface. This integrated pipeline:
- Consolidates multiple tools into one platform, eliminating the need to switch between systems
- Automates the complete workflow from raw VCF data to clinical interpretation
- Requires minimal technical expertise to operate effectively
Researchers and clinicians can now:
- Process VCF files to extract critical variant data
- Run SegmentNT analysis to identify genomic regions
- Query ClinVar for clinical significance
- Generate comprehensive reports
- Ask questions in natural language
All without ever leaving the interface or needing to reformat data between tools!
Unified Analysis Pipeline
- VCF file parsing and variant extraction
- SegmentNT neural network analysis for genomic region identification
- ClinVar database integration for clinical significance
- AI-powered report generation
- Natural language querying interface
- Advanced AI Capabilities
Powered by Google's Gemini 2.0 Flash model
- Retrieval Augmented Generation (RAG) for accurate variant analysis
- Grounded responses based on actual genomic data
- Structured report generation and formatting
- Interactive Interface
Intuitive Platform
- Real-time variant information display
- Chat-based interaction for queries
- Downloadable reports and analysis results
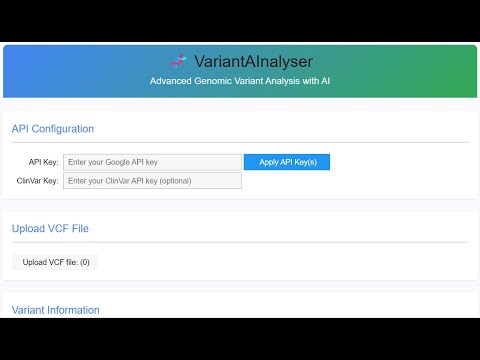
Before using VariantAInalyser, you'll need:
-
Google API key
- Generate it from AI Studio
- Add to the designated field in the interface
-
ClinVar API key (optional)
- Create a free NCBI account
- Navigate to "Account Settings"
- Generate an API key in the "API Key Management" section
- Add to the corresponding field in the interface
🗒️ Note: No worries if you don't have a ClinVar API key, you will still be able to use the VariantAInalyser interface. The ClinVar API key helps avoid rate limiting for multiple queries, but for typical use cases the system should work fine without it :)
- VCF File
You can download an example gzipped VCF folder containing multiple variants' VCF files from the official NCBI ClinVar webpage by clicking on this link: https://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh38/clinvar_papu.vcf.gz.
- Reference Genome File
To create the altered genome, a reference genome (i.e. with no variant) is needed. The majority of the variants present in the previously linked VCF folder are present in the Y chromosome. As such, I have uploaded the reference genome of chromosome Y as a FASTA file under the "Reference Genome" folder. However, if you require a different chromosome's reference genome, you can download it from "https://ftp.ensembl.org/pub/release-114/fasta/homo_sapiens/dna/". Once you download the reference genome's fasta file, upload it to your Google Colab notebook. The notebook directly links to the runtime files' location whenever it needs the reference genome so there is no need to make any changes in the code. However, to avoid having to reupload the files everytime you restart a runtime, you could also save the reference genome to your Google Drive, mount it to your Google Colab runtime and change the path to the relevant one in the prepare_altered_genome() method.
-
Clone the repository:
git clone https://github.com/yourusername/VariantAInalyser.git cd VariantAInalyser -
Open the notebook in Jupyter/Colab
-
Run all cells to initialise the interface
-
Enter your Google API key and Clinvar API key (this one is optional) into their corresponding boxes and click "Apply API Key/s"
-
Upload the VCF file containing genetic variants by clicking on the "Upload VCF file" button.
-
Start analysing the variants through the interactive interface!
🗒️ Note: Detailed example queries are included inside the notebook to ensure you get to experience all the features offered by the interface.
A demo of the VariantAInalyser interface in use can also be found below:
The pipeline generates several types of output:
- Detailed variant analysis reports
- SegmentNT probability plots
- Clinical significance assessments
- Comparative analyses
- Chat history logs
All outputs are automatically saved to the specified output directory.
Contributions are welcome! Please feel free to submit a pull request.
- Fork the repository
- Create your feature branch (
git checkout -b feature/yourfeature) - Commit your changes (
git commit -m 'Add some yourfeature') - Push to the branch (
git push origin feature/yourfeature) - Open a Pull Request
InstaDeep's SegmentNT for genomic analysis
Google's Gemini 2.0 Flash model for AI capabilities
NCBI ClinVar database for variant information